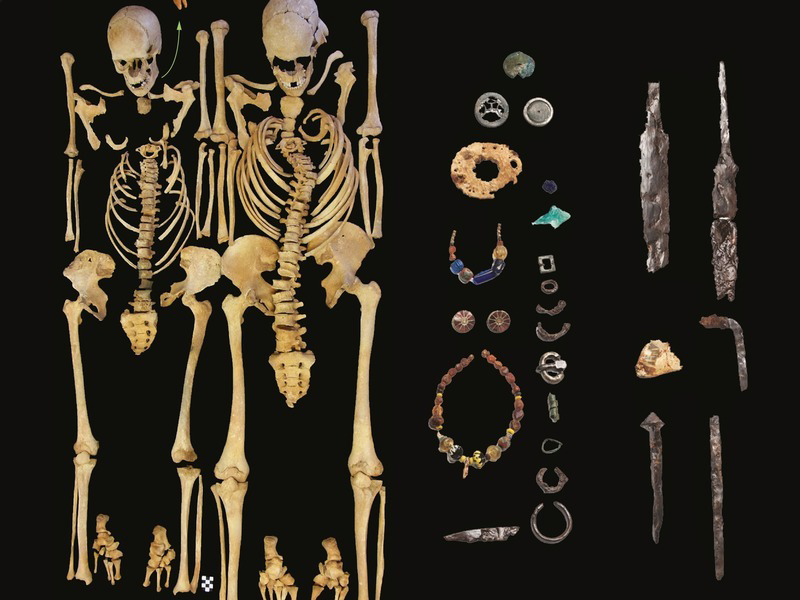
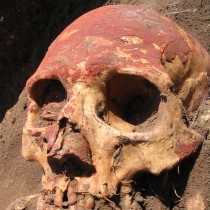
For the first time, scientists have succeeded to fully reconstruct a genome of the Justinianic Plague causative pathogen, Yersinia pestis, from a skeleton of a victim excavated 50 years ago in Altenerding, Southern Germany.
The research team that includes Johannes Krause, Alexander Herbig and Michal Feldman of the Max Planck Institute for the Science of Human History in Jena and Michaela Harback and colleagues of the Staatssammlung für Anthropologie und Paläoanatomie in Munich has produced a historical and high-quality reference data set that is an important resource for future analysis and has succeeded to add new insights into the evolution, adaptation and pathogenicity of a bacterium, which to this very day afflicts humankind.
Before the infamous Black Death, the first great plague epidemic was the Justinian plague, which, over the course of two centuries, wiped out up to an estimated 50 million of the world’s population throughout the Byzantine Empire – and may have helped speed the decline of the eastern Roman Empire. No one knows why it disappeared.
Recent molecular clues from ancient plague victims have suggested that plague may have been caused by the same bacterium, Yersinia pestis, which was responsible for the Black Death. But the geographic reach, mortality and impact of the Justinian pandemic are not fully known. Both information from ancient hosts and bacteria could shed light on the role of plague, which has afflicted mankind for more than 5,000 years.
Now, scientists based in Germany, including Michal Feldman, Alexander Herbig and Johannes Krause, of the Max Planck Institute for the Science of Human History in Jena and Michaela Harback and colleagues of the Staatssammlung für Anthropologie und Paläoanatomie in Munich have confirmed this by recovering the bacterial culprit from a sixth century skeletons found in Altenerding, an ancient southern German burial site near Munich. The Altenerding genome dates back to the beginning of the plague.
“Our research confirms that the Justinianic plague reached far beyond the historically documented affected region and provides new insights into the evolutionary history of Yersinia pestis, illustrating the potential of ancient genomic reconstructions to broaden our understanding of pathogen evolution and of historical events,” says researcher Michal Feldman.
The team has generated the first high-coverage genome of the bacterial agent responsible for the Justinian plague. In addition to revealing new insights in the molecular evolution of Yersinia pestis since the Byzantine times, the new sequence shows features that could not detected due to the limitations in the coverage of a draft genome previously reported, including 30 newly identified mutations and structural rearrangements unique to the Justinianic strain, as well as correcting 19 false positive mutations. Three are located in genes critical to plague virulence: nrdE, fadJ and pcp genes. Their data also suggested that the strain was more genetically diverse than previously thought.
“The fact that the archeological skeletons which gave these exciting insights were excavated over 50 years ago underscores the importance of maintaining well curated anthropological collections”, says Michaela Harbeck of State Collection of Anthropology and Palaeoanatomy Munich.
This new findings allow the authors to develop guidelines that could help improve the quality and authenticity of genomic data recovered from candidate ancient pathogens. And with plague classified as a re-emerging infectious disease in certain regions, an important historic, high-quality reference resource has been generated to offer insights into the evolutionary changes, adaptation and human impact of plague.